Critical Region
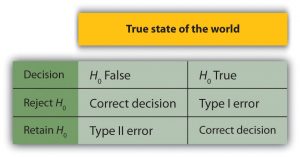
If the test statistic falls in this region under the normal distribution curve of the sample data the null hypothesis is rejected.
This region is depicted as the extreme value on either the left, right or both sides of the normal curve, that is near the tail.
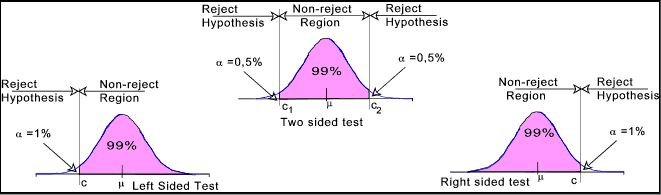
One-Tailed Null Hypothesis Testing
When the critical region ( i.e. null hypothesis rejection region ) is only one side of the normal distribution curve - either left tail or right tail it is called one-tailed null hypothesis.
It is also called directional hypothesis testing because the alternate hypothesis rejects the null hypothesis by comparing the test statistic value to be either greater than or less than the reference value specified in the null hypothesis.
For example:
H0: μ = 20 vs. H1: μ > 20
H0: μ = 200 vs. H1: μ < 300
Two-tailed Null Hypothesis Testing
When the critical region ( i.e. null hypothesis rejection region ) is on both sides of the normal distribution curve - left and right tail it is called two-tailed null hypothesis testing.
It is also called non-directional hypothesis testing because the alternate hypothesis rejects the null hypothesis if the test statistic value is not equal to the reference value specified in the null hypothesis - regardless of whether it is greater than or less than the specified value.
For example:
H0: μ = 850 vs. H1: μ≠ 850
One-Sample t.test
Understanding the Lung Capacity dataset
We load the tab separated data file which has the Lung Capacity dataset.
# load lung capacity data
# from text file with tab delimited columns
lungCapacity_df = read.csv("~/dataFiles/LungCapData.txt", header=T, sep="\t")
class(lungCapacity_df)
# [1] "data.frame"
# see the rows and columns in data frame
dim(lungCapacity_df)
# [1] 725 6
str(lungCapacity_df)
# 'data.frame': 725 obs. of 6 variables:
# $ LungCap : num 6.47 10.12 9.55 11.12 4.8 ...
# $ Age : int 6 18 16 14 5 11 8 11 15 11 ...
# $ Height : num 62.1 74.7 69.7 71 56.9 58.7 63.3 70.4 70.5 59.2 ...
# $ Smoke : Factor w/ 2 levels "no","yes": 1 2 1 1 1 1 1 1 1 1 ...
# $ Gender : Factor w/ 2 levels "female","male": 2 1 1 2 2 1 2 2 2 2 ...
# $ Caesarean: Factor w/ 2 levels "no","yes": 1 1 2 1 1 1 2 1 1 1 ...
names(lungCapacity_df)
# [1] "LungCap" "Age" "Height" "Smoke" "Gender" "Caesarean"
# see summary of lung capacity
summary(lungCapacity_df$LungCap)
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 0.507 6.150 8.000 7.863 9.800 14.680
Interpret the t.test result
Here test statistic is based on single statistic. For example mean of Lung Capacity for the given dataset.
Null Hypothesis: Mean of LungCap is greater than 8
H0: μ >= 8 vs. H1: μ < 8
# we test with 5 percent significance level
# that is, confidence interval 95 %
Test.one.sided.95 = t.test(lungCapacity_df$LungCap,
mu=8,
alt="less",
conf=0.95)
Test.one.sided.95
# One Sample t-test
# data: lungCapacity_df$LungCap
# t = -1.3842, df = 724, p-value = 0.08336
# alternative hypothesis: true mean is less than 8
# 95 percent confidence interval:
# -Inf 8.025974
# sample estimates:
# mean of x
# 7.863148
names(Test.one.sided.95)
# [1] "statistic" "parameter" "p.value" "conf.int" "estimate"
# [6] "null.value" "alternative" "method" "data.name"
# to reject or not to reject the null hypothesis
p = Test.one.sided.95$p.value
significance = 0.05
Result = ifelse( (p - significance)>0,
"NULL is True",
"Alternate is True: Reject Null" )
Result
# [1] "NULL is True"
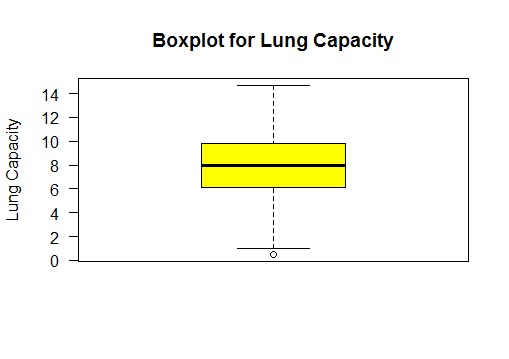
Boxplot the two sample populations
Group 1 : Lung Capacity of Non-Smokers
Group 2 : Lung Capacity of Smokers
It shows the lung capacity of non-smokers has higher variance than that of smokers.
# lung capacity for smokers and non-smokers
class(lungCapacity_df$LungCap)
#[1] "numeric"
class(lungCapacity_df$Smoke)
#[1] "factor"
summary(lungCapacity_df$LungCap)
# Min. 1st Qu. Median Mean 3rd Qu. Max.
# 0.507 6.150 8.000 7.863 9.800 14.680
summary(lungCapacity_df$Smoke)
# no yes
# 648 77
# box plot lungcapacity wrt to smoker - yes or no
boxplot(lungCapacity_df$LungCap~lungCapacity_df$Smoke)
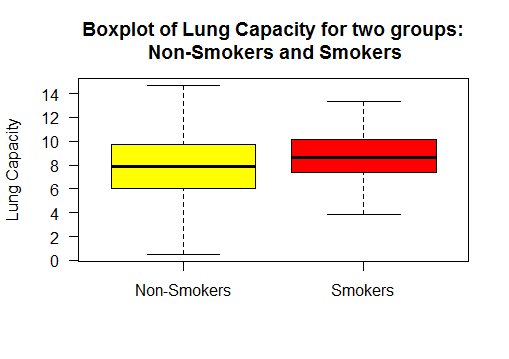
Paired t-test
Here the t-test is done on two populations that are dependent on each other.
We use the blood pressure dataset which records the blood pressure of patients before and after treatment.
Blood pressure before and after treatment
We compare the means of below two dependent populations:
μ1 - mean of blood pressure before treatment
μ2 - mean of blood pressure after treatment
Null Hypothesis: Two means are equal
Alternate Hypothesis : Two means are not equal, two-tailed testing
H0: μ1 = μ2 vs. H1: μ1 ≠ μ2
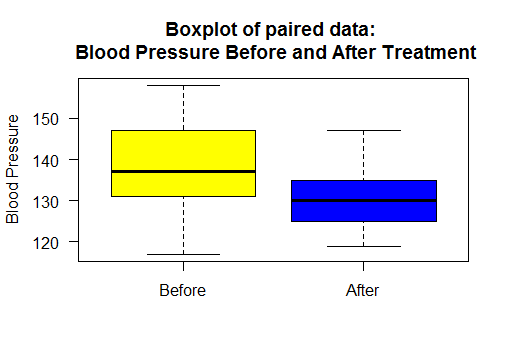
Boxplot and Scatter Plot for paired dataset
Boxplot of blood pressure before and after treatment.
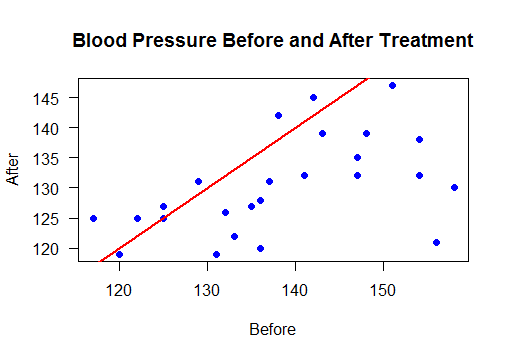
Scatter plot with regression line
Points below the line show reduced blood pressure after treatement
#boxplot
boxplot(bloodPressure$Before, bloodPressure$After,
names=c("Before","After"),
col=c("yellow","blue"),
main="Boxplot of paired data: \n Blood Pressure Before and After Treatment",
ylab="Blood Pressure", las=1)
#Scatter plot
plot(bloodPressure$Before, bloodPressure$After,
main="Blood Pressure Before and After Treatment",
xlab="Before", ylab="After",las=1,pch=19, col="blue")
abline(a=0, b=1, col="red",lwd=2)