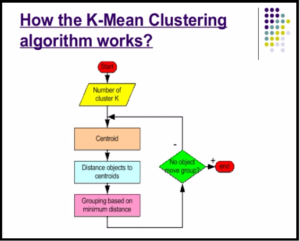
Programming Logic
Steps to cluster data points based on numerical features using KMeans clustering algorithm and compare the results with Hierarchical clustering.
Pre-requisite:
Understand the dataset for pre-processing that may be requierd for clustering algorithm.
Step 1:
Remove the categorical variable from the dataset.
Step 2:
Use k-means function for number of clusters.
Step 3:
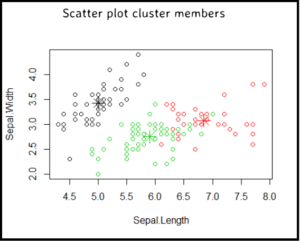
Scatter plot two dimensions colour coding data points per the k-means cluster members. Also plot the cluster centers and interpret the plot.
Step 4:
Compare the cluster members with actual values of the categorical variable.
Step 5:
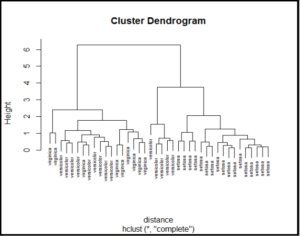
Now use Hierarchical clustering and plot Dendogram
Step 6:
Interpret Dendogram ad compare the with k-means scatter plot for cluster members.
Data pre-processing
We remove the categorical variable 'Species' from the dataset and use remaining four to identify the clusters using k-means clustering.
# now we remove the 'Species' column from the data
# this is to make sure the data is unlabeled
iris_numerical =iris[,-5]
str(iris_numerical)
#'data.frame': 150 obs. of 4 variables:
# $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
# $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
# $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
# $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
Scatter plot
Plot two dimensions colour coding the data points based on which KMeans cluster they belong to; and show the cluster centers.
# there are four dimensions to the data but we use only two
# plot Sepal length & width and colour as per cluster
# membership
plot(iris_numerical[c("Sepal.Length","Sepal.Width")],
col = kmeansClustering$cluster)
#plot cluster centers for the clusters
points(kmeansClustering$centers[,c("Sepal.Length", "Sepal.Width")], col =1:3, pch=8, cex=2)
Compare with actual values
Lets compare the cluster members with the actual total values for the categorical attribute.
#Lets compare the kmeans cluster with the Species classes
table(iris$Species, kmeansClustering$cluster)
# 1 2 3
# setosa 50 0 0
# versicolor 0 48 2
# virginica 0 14 36
# cluster setosa can be separated from other clusters
# clusters versicolor and virginica have a small degree of overlap
Plot Dendogram
See the Hierarchical clusters using dendogram.
# plot dendogram for hierarchical clustering
plot(iris_numerical_40_hc)
# plot(iris_numerical_40_hc, hang=-1, labels=iris$Species[index])
plot(iris_numerical_40_hc,
labels=iris$Species[index],
cex=0.6)