
Install required R packages
install.packages("ISLR")
install.packages("tree")
Understanding the data set to build prediction model
We will use Carseats dataset from ILSR library.
Target variable 'Sales' is a continuous variable, since we cannot do classification on continuous variable we create a new variable 'High' to indicate sales is high or not. We assign categorical values 'Yes' or 'No' to High based on condition that sales value is greater than the median sales value.
# check dimensions of data set
dim(Carseats)
#[1] 400 11
names(Carseats)
# [1] "Sales" "CompPrice" "Income" "Advertising"
# [5] "Population" "Price" "ShelveLoc" "Age"
# [9] "Education" "Urban" "US"
str(Carseats)
# 'data.frame': 400 obs. of 11 variables:
# $ Sales : num 9.5 11.22 10.06 7.4 4.15 ...
# $ CompPrice : num 138 111 113 117 141 124 115 136 132 132 ...
# $ Income : num 73 48 35 100 64 113 105 81 110 113 ...
# $ Advertising: num 11 16 10 4 3 13 0 15 0 0 ...
# $ Population : num 276 260 269 466 340 501 45 425 108 131 ...
# $ Price : num 120 83 80 97 128 72 108 120 124 124 ...
# $ ShelveLoc : Factor w/ 3 levels "Bad","Good","Medium": 1 2 3 3 1 1 3 2 3 3 ...
# $ Age : num 42 65 59 55 38 78 71 67 76 76 ...
# $ Education : num 17 10 12 14 13 16 15 10 10 17 ...
# $ Urban : Factor w/ 2 levels "No","Yes": 2 2 2 2 2 1 2 2 1 1 ...
# $ US : Factor w/ 2 levels "No","Yes": 2 2 2 2 1 2 1 2 1 2 ...
Set seed
To make result reproducible
set.seed(2)
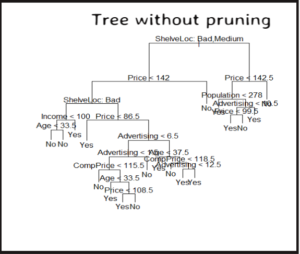
Fit the tree model
The model is built using training dataset and the predictor depends on all features.
tree_model = tree(High~., training_data)
Use the model to predict test data target
test_pred = predict(tree_model, testing_data, type="class")
Use Cross validation to improve accuracy of the tree model
The cross-validation function gives the size and corresponding deviance or error. 'Pruning' the tree model using the size with lowest deviance can help improve the accuracy
# To improve the accuracy we do cross validaion
# cross validation to check where to stop pruning
set.seed(3)
cv_tree = cv.tree(tree_model, FUN = prune.misclass)
names(cv_tree)
# [1] "size" "dev" "k" "method"
# size of prune trees
# dev - deviance or cv error rate
# plot the size and deviance to see where error is lowest
plot(cv_tree$size, cv_tree$dev, type="b",
xlab='Tree Size',
ylab='Error Rate',
main = 'Cross Validation: Error Vs Size')
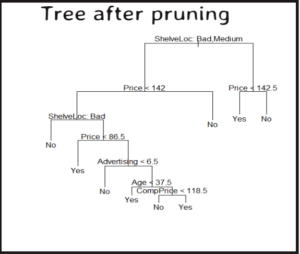
Prune tree model
Use best size from cross-validation plot for pruning to improve accuracy
# prune the model and plot
pruned_model = prune.misclass(tree_model, best = 9)
plot(pruned_model)
text(pruned_model, pretty = 0)
Measure accuracy of pruned model
And compare with the accuracy of tree model before pruning
#[1] 0.715
# test the accuracy of pruned model
tree_pred = predict(pruned_model, testing_data, type="class")
accuracy = mean(tree_pred == testing_High)
accuracy
# [1] 0.77
# Bit improvement compared to 0.71
